INCA enables robust metabolic tracer studies
A software tool for metabolic analysis developed by a Vanderbilt chemical engineer recently passed 1,000 total academic licenses and is the most licensed software on the university’s online licensing and e-commerce platform.
Additionally, it was the third highest revenue generator on the platform, VU e-Innovations, for 2020. About 20 total commercial licenses have been sold since INCA was released. In 2020, a total of 125 licenses, the bulk of them academic, were sold.
INCA, or Isotopomer Network Compartmental Analysis, was developed by Jamey Young and released in 2014 as part of the Metabolic Flux Analysis suite. Using INCA enables researchers to model data from metabolic tracer studies and quantify the flow of labeled atoms inside living cells. The information provided by INCA can help to better understand metabolic diseases or improve the productivity of cell-based bioprocesses. 
“We developed it to serve our own research needs,” said Young, professor of chemical and biomolecular engineering. “We were investigating the metabolism of tumor cells, how they use nutrients to grow and survive. We needed better software tools to make sense of the data we were collecting in the lab.”
INCA automates flux analysis of isotope tracer experiments, which has become an essential component of metabolic engineering research. The most common metabolic tracer is carbon-13, since nearly all metabolic reactions involve the transfer of carbon atoms from one compound into another.
An isotopic workhorse
Carbon-13 itself is rather mundane. And that is what makes it so valuable to chemical and biological research. Carbon-13 is chemically equivalent to the more abundant isotope carbon-12 and is metabolized by plants and animals in the same way. Normally, it accounts for only 1 percent of natural carbon, but researchers can procure nutrients like glucose that are enriched with carbon-13 at specific atom positions.

Researchers feed cells with the labeled nutrient, and the carbon-13 acts as a tag that can be traced through metabolic reactions and detected with mass spectrometry.
“Carbon metabolism is central to all cells,” Young said. “We are able to trace the major metabolic pathways of bacteria, plants, or animals using carbon-13. The fact that it is not radioactive makes it safe and easy to use in the lab.”
Similar to the way Google uses cell phone data to trace the flow of cars on the highway and produce a traffic map, analysis of carbon-13 labeling data with INCA can provide a “flux map” describing the flow of atoms through metabolic pathways inside of cells.
“Most of our experiments are designed to compare those traffic flows under different conditions, to see how cells behave under normal conditions versus in response to a disease, drug treatment, or a genetic change in the cells’ DNA,” Young said.
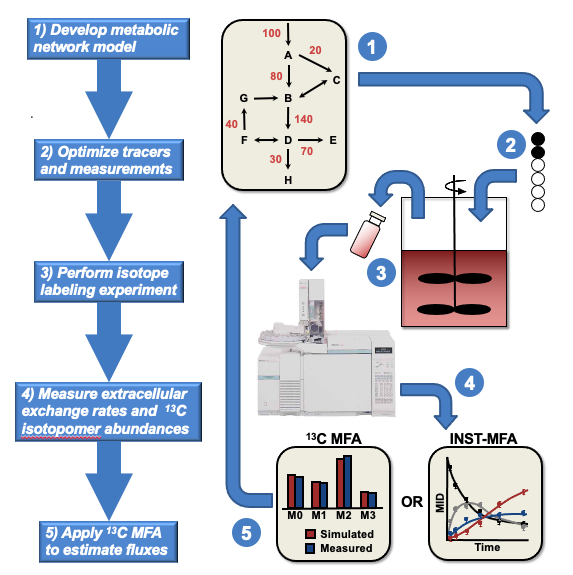
He plans to release a major update—the first since 2014—to INCA later this year. INCA, which is MATLAB-based, is further distinguished as the first publicly available software package that can analyze both steady-state and dynamic labeling experiments. The latter are especially relevant to photosynthetic organisms and cell cultures that metabolize the tracer very slowly.
The Young lab has used INCA to achieve several breakthroughs, including the first comprehensive carbon-13 flux studies of photosynthetic bacteria and plant leaves. Young also co-founded Metalytics, Inc., which uses INCA to provide metabolic flux analysis services to the biotech industry.
Wide use in industry and academia
Many companies and academic labs are interested in using INCA to speed up the production of biochemicals or drugs made from genetically engineered cells. By pinpointing traffic bottlenecks and inefficiencies in the metabolism of the host cells, researchers can add or remove genes to create detours or block paths that lead in the wrong direction. Biomedical researchers also use INCA to understand how metabolic fluxes are misdirected due to a disease, which can lead to new treatments or diagnostics that target the basic cellular mechanisms of the illness.
Experiments that measure carbon-13 enrichment in multiple metabolites produce large, complex data sets. INCA allows users to rigorously model the data and quantify metabolic rates, Young said. “You can make some sense of the data without using software, but INCA enables you to extract more information from it and to integrate many different measurements into a single, comprehensive flux map.”
A demonstration video of INCA is available here.
INCA and other applications for metabolic flux analysis are categorized as metabolomics software. Young is part of a National Institutes of Health working group that is developing standards for such software, including best practices for design and documentation to make the software user-friendly. Guidelines were published in January 2021.
VU e-Innovations is CTTC’s online licensing and e-commerce platform to commercialize innovative software, courseware and other specialized products developed exclusively at Vanderbilt. The platform automates the licensing of certain digital or downloadable IP assets to interested users, who can access such programs by agreeing to the terms of an end user license agreement, and, in the case of commercial licenses, paying the licensing fee. Commercial entities pay an annual fee for each product; academic licenses are free.
“Software and digital tools such as INCA and the Metabolic Flux Analysis suite developed by the Young Lab are an important component of Vanderbilt’s digital technologies portfolio. They support discovery and innovation far beyond Vanderbilt,” said Masood Ahammed Machingal, manager, digital technology licensing
Building on the successes of non-patented intellectual property assets like INCA, CTTC is expanding this platform (VU e-Innovations 2.0 is in the works) and is currently accepting software, courseware, surveys and other digital copyrightable materials from Vanderbilt researchers for release via VU e-Innovations.